History of the Human Genome Assembly
22 years, 3,117,275,501 bases and 0 gaps later
Round numbers are always false.
— Samuel Johnson

contents
- 1 · Glossary of terms
- 2 · Human genome assembly through the years
- 3 · Human genome assembly — the last step
- 4 · Creating the Scientific American graphic
- 4.1 · Assembly alignments to CHM13v2
- 4.2 · CHM13v2 alignment coverage
- 4.3 · Assembly history
- 4.4 · Incremental assembly history
- 4.5 · Cumulative assembly history
- 5 · Assembly history in visual form
First, a few terms to know. For more definitions, see the NIH glossary of genomic and genetic terms.
base — one of the the nucleotides A, T, G or C (in genomic sequence) and a fundamental unit of genetic information
chromosome — a threadlike structure comprising a single molecule of DNA along with proteins that create a top-level package of genomic information and serve to carry it from cell to cell
mitocondrial chromosome — a very short (e.g. human chrM is 16,569 bases) maternally-inherited circular chromosome found inside mitochondria that encode proteins involved in oxidative phosphorylation system
sequence read — an individual sequence fragment read out by a DNA sequencer, which may vary in length from 10's to 1000's of bases
contig — continuous stretches of sequence containing without gaps (i.e. containing only A, C, G, or T bases)
scaffold — an ordered set of contigs separated by gaps that are created using additional information about the relative position and orientation of the contigs
gap — a stretch of N's in a sequence representing a gap in the sequence due to lack of coverage whose length may be set to certain fixed values to represent the reason for the gap (e.g. contig gap).
assembly — an ordered and oriented set of nucleotide sequences (usually scaffolds)
fasta file — a plain-text file format for storing sequence information
Before I describe the method used to generate the graphic, let's look at some details of how the human genome assembly changed over the years, in the context of sizes of sequenced regions and gaps.
In the table below, I show the total sequence size of each assembly from hg1 (May 2000) through hg38 (Dec 2013) (download hg assemblies) to the gapless CHM13v2 telomere-to-telomere (T2T) assembly (Mar 2022). Here, the sequence size reflects the number of bases in the assemblies of chromosomes 1–22, X, and Y and excludes the mitochondrial chromosome and any unanchored, alternate or random content.
assembly year month seq +seq seq/CHM gap_n gap_size hg1 2000 May 2,429,568,807 77.94% 756,351 386,046,812 hg2 2000 June 2,407,220,830 -22,347,977 77.22% 503,500 590,322,429 hg4 2000 Sep 2,592,759,797 185,538,967 83.17% 332,807 508,661,915 hg5 2000 Oct 2,662,673,274 69,913,477 85.42% 338,786 647,331,541 hg6 2000 Dec 2,679,206,778 16,533,504 85.95% 296,300 571,716,303 hg7 2001 Apr 2,733,032,147 53,825,369 87.67% 265,972 524,112,663 hg8 2001 Aug 2,849,390,173 116,358,026 91.41% 232,126 529,686,227 hg10 2001 Dec 2,795,152,898 -54,237,275 89.67% 124,985 350,152,047 hg11 2002 Apr 2,824,723,360 29,570,462 90.62% 89,709 299,451,070 hg12 2002 June 2,810,120,127 -14,603,233 90.15% 70,360 231,723,944 hg13 2002 Nov 2,843,602,073 33,481,946 91.22% 69,862 207,028,754 hg15 2003 Apr 2,832,183,299 -11,418,774 90.85% 507 238,337,817 hg16 2003 July 2,843,413,503 11,230,204 91.21% 412 226,714,556 hg17 2004 May 2,851,330,913 7,917,410 91.47% 345 225,450,974 hg18 2006 Mar 2,858,012,806 6,681,893 91.68% 333 222,406,674 hg19 2009 Feb 2,861,327,131 3,314,325 91.79% 362 234,350,281 hg38 2013 Dec 2,937,639,113 76,311,982 94.24% 1,001 150,630,719 CHM13v2 2022 Mar 3,117,275,501 179,636,388 100.00% 0 0
The public open-source assemblies were indexed by hgN from hg1 (May 2000) to hg38 (Dec 2013). Differences in the index value do not necessarily represent how much of the assembly changed and some indexes were skipped. Notably, although hg3 (July 2000) is reported to exist, I have been unable to locate its sequence files. Apparently, nobody can find the sequence files for hg3!
In the first assembly, hg1 (May 2000), chromosomes were reported in individual contigs. A contig is a contiguous sequence assembly and may have gaps. Chromosomes 1–22, X and Y comprised 1,620 contigs — for example, chromosome 1 was reported in 149 contigs. The number of gaps and gap size for hg1 in the table above does not reflect the gaps between contigs. In subsequent assemblies, these contigs were concatenated (with a contig-level gap as a separator) into a chromosome-level assembly.
One way to understand the pace of development of the human genome assembly is to look over the years at (a) how much sequence was added to each chromosome and (b) how many gaps (regions of unknown sequence) were closed.
For example, from 2000 to 2022 the amount of sequence in chromosome 1 changed from 210,989,981 to 248,387,328 bases, while the number of gaps in chromosome 1 dropped from 19,758 to zero.
Early in the assembly's history, some chromosome sizes decreased as misassembled regions were moved to their correct chromosome. More recently, chromosome sizes increased as gaps were filled in and difficult-to-sequence regions were completed.
The interesting consequence to this is that the order of chromosomes by their label no longer reflects their order by size. For example, chr9, chr11, chr20 and chr22 are all larger than their preceeding counterparts chr8, chr10, chr19, chr21, respectively.
In the table below, I show how the amount of sequence in each chromosome's assembly changed through the years — the most recent assembly within a given year is reported. Years in which the chromosome size increased by more than 2%, 5% or 10% are shown in light to dark green. Where size decreased by 1%, 2.5% or 4%, lines are shown in light to dark pink.
The values here represent the amount of known sequence in a chromosome — its total length would be this value plus the total size of gaps in the chromosome. For the CHM13v2 assembly, since there are no gaps, this value is equal to the length of the chromosome assembly.
Assembly names preceeded by * are the last assemblies for their given year and form the basis for the year's representation in the graphic. These were
hg6 (Dec 2000),
hg10 (Dec 2001),
hg13 (Nov 2002),
hg16 (July 2003),
hg17 (May 2004),
hg18 (Mar 2006),
hg19 (Feb 2009),
hg38 (Dec 2013).
len (bases) Δ(%) assembly year ----- chr1 ----- ----- chr2 ----- ----- chr3 ----- ----- chr4 ----- hg1 2000 168,913,425 196,386,279 163,542,702 144,098,731 hg2 2000 183,709,012 8.1 194,383,237 -1.0 165,584,260 1.2 133,549,012 -7.9 hg4 2000 201,521,296 8.8 214,586,282 9.4 176,970,320 6.4 148,983,448 10.4 hg5 2000 209,982,297 4.0 220,854,774 2.8 185,825,750 4.8 167,597,939 11.1 *hg6 2000 210,989,981 0.5 221,356,626 0.2 185,824,536 -0.0 169,058,312 0.9 hg7 2001 222,977,559 5.4 221,725,538 0.2 184,829,848 -0.5 171,864,472 1.6 hg8 2001 234,799,493 5.0 225,165,963 1.5 194,242,316 4.8 176,201,768 2.5 *hg10 2001 227,673,339 -3.1 226,391,327 0.5 184,536,607 -5.3 172,328,593 -2.2 hg11 2002 221,443,782 -2.8 238,221,181 5.0 187,464,974 1.6 187,759,065 8.2 hg12 2002 220,881,717 -0.3 236,107,886 -0.9 189,490,410 1.1 187,561,753 -0.1 *hg13 2002 221,229,757 0.2 237,456,869 0.6 194,222,908 2.4 187,608,462 0.0 hg15 2003 218,712,898 -1.2 237,043,673 -0.2 193,607,218 -0.3 186,580,523 -0.6 *hg16 2003 221,562,941 1.3 237,541,603 0.2 194,473,776 0.4 186,841,959 0.1 *hg17 2004 222,827,847 0.6 237,503,374 -0.0 194,635,735 0.1 187,161,218 0.2 *hg18 2006 224,999,719 1.0 237,709,794 0.1 194,704,822 0.0 187,297,063 0.1 *hg19 2009 225,280,621 0.1 238,204,518 0.2 194,797,135 0.0 187,661,676 0.2 *hg38 2013 230,481,012 2.3 240,548,228 1.0 198,100,135 1.7 189,752,667 1.1 CHM13v2 2022 248,387,328 7.2 242,696,752 0.9 201,105,948 1.5 193,574,945 2.0 assembly year ----- chr5 ----- ----- chr6 ----- ----- chr7 ----- ----- chr8 ----- hg1 2000 164,809,671 146,037,284 126,787,148 112,574,678 hg2 2000 166,925,488 1.3 149,388,392 2.2 121,050,404 -4.7 110,307,747 -2.1 hg4 2000 163,940,667 -1.8 159,837,923 6.5 139,317,378 13.1 120,650,901 8.6 hg5 2000 167,994,833 2.4 158,085,470 -1.1 145,591,522 4.3 124,248,692 2.9 *hg6 2000 166,043,302 -1.2 158,660,857 0.4 147,591,730 1.4 124,912,316 0.5 hg7 2001 164,878,747 -0.7 165,881,551 4.4 147,038,945 -0.4 129,385,700 3.5 hg8 2001 172,166,415 4.2 176,828,634 6.2 156,866,542 6.3 137,308,140 5.8 *hg10 2001 169,841,985 -1.4 172,818,856 -2.3 156,983,175 0.1 133,762,905 -2.7 hg11 2002 181,189,438 6.3 176,229,400 1.9 155,514,763 -0.9 140,091,045 4.5 hg12 2002 176,708,043 -2.5 166,004,516 -6.2 153,794,993 -1.1 139,191,073 -0.6 *hg13 2002 177,638,514 0.5 166,765,675 0.5 153,794,793 -0.0 142,314,151 2.2 hg15 2003 177,524,972 -0.1 166,880,540 0.1 154,546,299 0.5 141,694,337 -0.4 *hg16 2003 177,552,822 0.0 167,256,575 0.2 154,676,518 0.1 142,347,919 0.5 *hg17 2004 177,702,766 0.1 167,317,698 0.0 154,759,139 0.1 142,612,826 0.2 *hg18 2006 177,702,766 0.0 167,273,991 -0.0 154,952,424 0.1 142,612,826 0.0 *hg19 2009 177,695,260 -0.0 167,395,066 0.1 155,353,663 0.3 142,888,922 0.2 *hg38 2013 181,265,378 2.0 170,078,522 1.6 158,970,131 2.3 144,768,136 1.3 CHM13v2 2022 182,045,439 0.4 172,126,628 1.2 160,567,428 1.0 146,259,331 1.0 assembly year ----- chr9 ----- ----- chr10----- ----- chr11----- ----- chr12----- hg1 2000 96,580,761 112,082,289 114,442,972 117,488,755 hg2 2000 101,613,999 5.0 119,941,602 6.6 119,010,047 3.8 113,377,946 -3.6 hg4 2000 103,119,589 1.5 123,122,208 2.6 127,294,940 6.5 123,024,623 7.8 hg5 2000 106,904,027 3.5 126,683,514 2.8 128,591,838 1.0 124,426,716 1.1 *hg6 2000 109,839,313 2.7 127,014,353 0.3 129,303,236 0.6 125,990,086 1.2 hg7 2001 111,915,677 1.9 131,169,312 3.2 133,764,924 3.3 129,714,762 2.9 hg8 2001 119,660,584 6.5 135,125,817 2.9 136,554,494 2.0 134,784,902 3.8 *hg10 2001 115,158,754 -3.9 132,754,952 -1.8 133,828,821 -2.0 129,564,139 -4.0 hg11 2002 112,776,971 -2.1 132,211,963 -0.4 127,267,213 -5.2 124,711,282 -3.9 hg12 2002 115,238,209 2.1 130,722,965 -1.1 130,025,188 2.1 126,156,683 1.1 *hg13 2002 116,960,681 1.5 130,943,927 0.2 132,130,624 1.6 128,265,502 1.6 hg15 2003 115,187,714 -1.5 130,710,865 -0.2 130,709,420 -1.1 129,328,332 0.8 *hg16 2003 115,624,042 0.4 131,173,206 0.4 130,908,854 0.2 129,826,277 0.4 *hg17 2004 117,781,268 1.8 131,613,619 0.3 131,130,753 0.2 130,259,309 0.3 *hg18 2006 120,143,252 2.0 131,624,728 0.0 131,130,753 0.0 130,303,032 0.0 *hg19 2009 120,143,431 0.0 131,314,738 -0.2 131,129,516 -0.0 130,481,393 0.1 *hg38 2013 121,790,550 1.4 133,262,962 1.5 134,533,742 2.5 133,137,816 2.0 CHM13v2 2022 150,617,247 19.1 134,758,134 1.1 135,127,769 0.4 133,324,548 0.1 assembly year ----- chr13----- ----- chr14----- ----- chr15----- ----- chr16----- hg1 2000 85,645,372 79,343,804 67,826,689 74,917,520 hg2 2000 87,721,609 2.4 81,516,980 2.7 66,300,573 -2.3 71,700,752 -4.5 hg4 2000 90,313,480 2.9 88,301,397 7.7 72,271,491 8.3 65,176,734 -10. hg5 2000 92,930,944 2.8 86,324,201 -2.3 73,349,533 1.5 71,375,964 8.7 *hg6 2000 93,269,031 0.4 86,368,090 0.1 74,363,214 1.4 73,084,034 2.3 hg7 2001 96,607,817 3.5 87,531,345 1.3 75,875,723 2.0 74,218,216 1.5 hg8 2001 97,944,675 1.4 88,761,063 1.4 80,448,614 5.7 81,170,232 8.6 *hg10 2001 97,023,436 -0.9 88,460,447 -0.3 77,195,556 -4.2 76,728,727 -5.8 hg11 2002 96,006,001 -1.1 87,931,086 -0.6 79,566,986 3.0 77,742,614 1.3 hg12 2002 95,206,001 -0.8 87,163,614 -0.9 79,644,830 0.1 77,609,164 -0.2 *hg13 2002 95,228,136 0.0 88,115,227 1.1 82,886,735 3.9 80,565,380 3.7 hg15 2003 95,511,656 0.3 87,191,216 -1.1 81,117,055 -2.2 79,890,791 -0.8 *hg16 2003 95,559,980 0.1 87,191,216 0.0 81,259,656 0.2 79,932,429 0.1 *hg17 2004 95,559,980 0.0 88,290,585 1.2 81,341,915 0.1 78,884,752 -1.3 *hg18 2006 95,559,980 0.0 88,290,585 0.0 81,341,915 0.0 78,884,752 0.0 *hg19 2009 95,589,878 0.0 88,289,540 -0.0 81,694,766 0.4 78,884,753 0.0 *hg38 2013 97,983,125 2.4 90,568,149 2.5 84,641,325 3.5 81,805,943 3.6 CHM13v2 2022 113,566,686 13.7 101,161,492 10.5 99,753,195 15.1 96,330,374 15.1 assembly year ----- chr17----- ----- chr18----- ----- chr19----- ----- chr20----- hg1 2000 67,776,776 70,211,394 49,532,089 65,503,175 hg2 2000 67,302,722 -0.7 70,585,267 0.5 48,893,614 -1.3 59,753,369 -9.6 hg4 2000 69,520,272 3.2 72,643,538 2.8 50,903,878 3.9 63,588,720 6.0 hg5 2000 72,210,252 3.7 72,442,291 -0.3 55,122,885 7.7 60,221,228 -5.6 *hg6 2000 73,859,773 2.2 72,035,922 -0.6 53,798,088 -2.5 61,555,802 2.2 hg7 2001 75,361,783 2.0 72,931,330 1.2 54,929,280 2.1 59,153,863 -4.1 hg8 2001 77,552,140 2.8 77,219,105 5.6 59,871,347 8.3 59,153,863 0.0 *hg10 2001 76,771,996 -1.0 74,351,766 -3.9 58,362,679 -2.6 59,153,863 0.0 hg11 2002 75,590,181 -1.6 74,435,887 0.1 56,327,192 -3.6 59,387,171 0.4 hg12 2002 75,718,615 0.2 73,033,376 -1.9 56,150,739 -0.3 59,422,997 0.1 *hg13 2002 79,580,998 4.9 74,601,365 2.1 56,230,936 0.1 59,424,940 0.0 hg15 2003 77,480,855 -2.7 74,534,531 -0.1 55,780,860 -0.8 59,424,990 0.0 *hg16 2003 77,677,744 0.3 74,654,041 0.2 55,785,651 0.0 59,424,990 0.0 *hg17 2004 77,800,220 0.2 74,656,155 0.0 55,785,651 0.0 59,505,253 0.1 *hg18 2006 77,800,220 0.0 74,656,155 0.0 55,785,651 0.0 59,505,253 0.0 *hg19 2009 77,795,210 -0.0 74,657,229 0.0 55,808,983 0.0 59,505,520 0.0 *hg38 2013 82,920,204 6.2 80,089,605 6.8 58,440,758 4.5 63,944,257 6.9 CHM13v2 2022 84,276,897 1.6 80,542,538 0.6 61,707,364 5.3 66,210,255 3.4 assembly year ----- chr21----- ----- chr22----- ----- chrX ----- ----- chrY ----- hg1 2000 38,719,201 34,749,742 111,552,961 20,045,389 hg2 2000 33,923,840 -14. 33,475,975 -3.8 87,142,038 -28. 20,062,945 0.1 hg4 2000 33,823,978 -0.3 33,785,749 0.9 128,815,270 32.4 21,245,715 5.6 hg5 2000 33,823,978 0.0 33,785,749 0.0 122,546,667 -5.1 21,752,210 2.3 *hg6 2000 33,823,978 0.0 33,785,749 0.0 124,872,836 1.9 21,805,613 0.2 hg7 2001 33,823,978 0.0 33,785,749 0.0 131,614,405 5.1 22,051,623 1.1 hg8 2001 33,823,978 0.0 33,821,688 0.1 137,258,174 4.1 22,660,226 2.7 *hg10 2001 33,827,908 0.0 33,821,688 0.0 141,143,154 2.8 22,668,225 0.0 hg11 2002 33,829,102 0.0 33,821,688 0.0 142,536,150 1.0 22,668,225 0.0 hg12 2002 33,834,561 0.0 33,821,688 0.0 143,962,881 1.0 22,668,225 0.0 *hg13 2002 33,917,713 0.2 33,821,688 0.0 147,236,866 2.2 22,660,226 -0.0 hg15 2003 33,924,742 0.0 34,352,051 1.5 147,686,664 0.3 22,761,097 0.4 *hg16 2003 33,924,307 -0.0 34,352,051 0.0 149,215,391 1.0 24,649,555 7.7 *hg17 2004 34,170,106 0.7 34,764,789 1.2 150,394,264 0.8 24,871,691 0.9 *hg18 2006 34,170,106 0.0 34,851,311 0.2 151,058,754 0.4 25,652,954 3.0 *hg19 2009 35,106,642 2.7 34,894,545 0.1 151,100,560 0.0 25,653,566 0.0 *hg38 2013 40,088,619 12.4 39,159,777 10.9 154,893,029 2.4 26,415,043 2.9 CHM13v2 2022 45,090,682 11.1 51,324,926 23.7 154,259,566 -0.4 62,460,029 57.7
As the human genome assembly matured from 2000 to 2022, the number of gaps in each chromosome initially dropped rapidly (from thousands to tens). In 2013 gaps increased slightly as more fine structure in the genome was discovered (e.g. a previous gap was filled in with gapped sequence).
HOW IS THE SIZE OF A GAP DETERMINED Precise size of certain gaps can be determined from information about distance between known sequences. When the size of the gap was not known, it was set to 100 bases following tradition. Other sizes, such as 1kb, 10kb and 50kb signalled more information about the nature of the gap, such as a contig gap, short arm gap, telomere gap, and so on.
The table below shows the number of gaps and their total size. As before, the last assembly in a given year is marked by *.
gap_n gap_size assembly year ------ chr1 ----- ------ chr2 ----- ----- chr3 ------ ------ chr4 ----- hg1 2000 34,616 30,489,476 38,379 31,527,834 84,260 30,313,573 32,913 29,049,049 hg2 2000 34,492 54,563,791 36,285 43,413,879 55,821 46,275,304 28,840 38,331,234 hg4 2000 21,036 41,143,057 24,754 36,101,572 35,994 41,066,838 17,851 35,305,214 hg5 2000 22,497 72,211,367 24,769 32,401,809 36,836 41,698,828 19,250 34,730,408 *hg6 2000 19,758 65,030,485 22,153 26,651,434 32,672 33,172,331 18,464 31,116,843 hg7 2001 15,016 55,714,365 17,631 25,388,886 30,856 30,827,190 18,062 28,884,701 hg8 2001 12,640 52,234,435 13,443 28,448,646 33,344 33,346,591 17,268 33,697,964 *hg10 2001 4,941 28,748,886 5,803 14,877,744 15,626 20,316,754 7,923 19,059,649 hg11 2002 3,245 24,489,948 2,010 8,536,007 11,060 17,241,853 3,248 9,196,379 hg12 2002 2,258 25,992,617 999 4,573,714 7,601 5,417,726 1,794 4,457,625 *hg13 2002 1,117 23,029,017 286 4,539,918 3,139 5,335,436 1,167 4,060,816 hg15 2003 104 26,491,000 29 6,271,355 32 5,804,513 17 5,030,000 *hg16 2003 65 24,565,000 30 6,074,355 12 4,870,274 17 4,890,000 *hg17 2004 37 22,695,000 28 5,514,855 10 4,870,005 15 4,250,000 *hg18 2006 39 22,250,000 25 5,241,355 10 4,797,005 14 3,976,000 *hg19 2009 39 23,970,000 24 4,994,855 9 3,225,295 12 3,492,600 *hg38 2013 168 18,475,410 35 1,645,301 29 195,424 18 461,888 CHM13v2 2022 0 0 0 0 0 0 0 0 assembly year ------ chr5 ----- ------ chr6 ----- ----- chr7 ------ ------ chr8 ----- hg1 2000 34,513 23,854,137 21,616 30,396,288 15,613 19,560,492 32,873 25,153,250 hg2 2000 34,359 37,141,477 20,393 33,662,304 11,347 27,333,082 21,668 29,107,452 hg4 2000 24,947 35,689,483 10,030 29,441,631 7,951 23,195,378 15,336 28,326,522 hg5 2000 26,341 35,090,699 9,000 24,329,772 7,607 21,032,384 16,211 28,527,729 *hg6 2000 23,369 31,341,535 5,793 22,664,501 6,455 18,655,717 13,587 22,822,038 hg7 2001 19,815 30,327,389 4,053 17,654,602 4,713 16,785,730 12,045 20,156,303 hg8 2001 19,187 29,103,247 3,276 15,233,714 5,076 15,679,731 10,899 26,912,672 *hg10 2001 8,428 14,999,700 955 5,498,715 1,507 6,827,041 10,150 11,967,677 hg11 2002 5,240 8,381,121 1,434 6,596,115 405 4,942,264 8,634 8,342,191 hg12 2002 3,586 4,258,357 21 4,305,001 13 3,637,800 10,083 4,683,249 *hg13 2002 1,308 4,124,045 16 3,905,001 13 3,637,800 12,833 3,990,968 hg15 2003 17 3,442,323 13 3,860,001 12 3,885,000 17 4,214,401 *hg16 2003 13 3,482,100 11 3,658,001 12 3,869,000 13 3,960,900 *hg17 2004 7 3,155,100 11 3,658,001 12 3,869,000 10 3,662,000 *hg18 2006 7 3,155,100 11 3,626,001 12 3,869,000 10 3,662,000 *hg19 2009 7 3,220,000 11 3,720,001 17 3,785,000 9 3,475,100 *hg38 2013 37 272,881 16 727,457 20 375,842 12 370,500 CHM13v2 2022 0 0 0 0 0 0 0 0 assembly year ------ chr9 ----- ------ chr10----- ----- chr11------ ------ chr12----- hg1 2000 25,171 14,732,314 32,064 19,073,156 121.7k 17,165,137 63,425 17,941,551 hg2 2000 25,115 21,450,767 32,131 22,379,421 21,177 25,077,447 33,437 26,735,971 hg4 2000 13,875 14,743,038 18,569 17,194,557 15,052 21,488,532 22,972 20,761,832 hg5 2000 12,363 35,367,417 20,421 18,905,774 14,539 22,191,715 22,107 19,855,773 *hg6 2000 10,060 31,423,962 17,009 16,285,393 12,737 18,204,889 17,732 18,732,591 hg7 2001 12,655 30,026,140 14,058 13,620,198 11,255 16,809,265 12,724 16,628,828 hg8 2001 11,017 33,199,108 7,646 11,726,572 9,229 18,010,260 9,590 19,291,470 *hg10 2001 7,882 17,868,638 2,293 9,339,871 6,248 7,605,834 5,013 10,068,038 hg11 2002 886 16,342,266 1,562 6,297,316 5,592 6,811,051 4,133 9,469,427 hg12 2002 880 17,200,547 420 3,693,785 3,140 7,417,357 4,169 5,143,889 *hg13 2002 328 15,916,433 223 3,703,975 7,300 4,390,398 1,993 5,116,887 hg15 2003 50 19,318,105 35 4,770,009 11 4,269,364 15 4,136,102 *hg16 2003 49 20,748,003 31 3,864,009 10 3,574,100 13 2,252,102 *hg17 2004 41 20,648,000 27 3,800,009 9 3,321,631 14 2,190,502 *hg18 2006 38 20,130,000 26 3,750,009 9 3,321,631 13 2,046,502 *hg19 2009 40 21,070,000 25 4,220,009 9 3,877,000 13 3,370,502 *hg38 2013 46 16,604,167 80 534,460 17 552,880 30 137,493 CHM13v2 2022 0 0 0 0 0 0 0 0 assembly year ------ chr13----- ------ chr14----- ----- chr15------ ------ chr16----- hg1 2000 22,350 12,581,994 11,292 5,951,248 22,036 10,756,252 33,927 9,641,803 hg2 2000 21,859 16,427,024 10,389 11,172,142 20,436 17,463,304 31,479 19,997,227 hg4 2000 10,373 11,559,689 4,084 10,153,243 13,887 13,424,263 31,575 19,282,293 hg5 2000 10,800 26,813,954 3,010 20,629,120 11,631 28,030,988 36,487 32,922,367 *hg6 2000 6,885 23,947,619 1,624 19,354,512 9,734 25,699,200 36,749 27,059,711 hg7 2001 3,687 22,178,932 1,455 18,707,124 8,079 24,832,783 43,288 27,983,727 hg8 2001 1,580 20,795,461 1,208 17,899,680 7,363 27,584,051 39,960 27,445,559 *hg10 2001 304 18,067,447 365 18,008,690 5,346 21,862,800 18,525 17,071,880 hg11 2002 117 17,608,619 194 17,456,551 5,750 20,453,173 15,980 15,505,131 hg12 2002 12 18,240,103 92 17,161,294 5,689 19,572,525 2,975 4,062,421 *hg13 2002 7 16,070,000 191 13,103,018 12,684 13,711,627 976 10,646,501 hg15 2003 7 18,640,000 2 18,120,000 11 18,997,000 17 10,105,208 *hg16 2003 6 17,483,000 2 18,120,000 11 18,997,000 15 10,109,503 *hg17 2004 6 18,583,000 2 18,078,000 11 18,997,000 6 9,942,502 *hg18 2006 6 18,583,000 2 18,078,000 11 18,997,000 6 9,942,502 *hg19 2009 6 19,580,000 2 19,060,000 12 20,836,626 6 11,470,000 *hg38 2013 23 16,381,203 25 16,475,569 19 17,349,864 22 8,532,402 CHM13v2 2022 0 0 0 0 0 0 0 0 assembly year ------ chr17----- ------ chr18----- ----- chr19------ ------ chr20----- hg1 2000 20,451 8,372,294 71,970 9,356,951 7,675 7,830,890 11,919 7,629,675 hg2 2000 21,778 16,924,590 12,853 18,463,986 8,615 20,861,435 7,973 8,378,942 hg4 2000 17,520 15,918,208 9,136 13,345,396 4,740 18,369,268 3,070 8,565,162 hg5 2000 19,692 17,294,301 9,443 14,235,257 5,889 19,839,960 983 6,446,777 *hg6 2000 19,720 15,321,291 8,771 11,770,782 4,488 17,209,846 967 5,641,164 hg7 2001 19,263 13,057,941 8,464 10,947,687 4,061 17,436,332 8 5,709,984 hg8 2001 14,724 12,324,351 7,656 12,987,228 3,970 12,430,032 8 5,709,984 *hg10 2001 12,387 7,091,707 7,810 7,448,235 1,012 18,563,646 9 3,809,988 hg11 2002 10,024 5,522,111 7,435 6,426,323 1,002 16,673,579 6 3,455,826 hg12 2002 17,806 4,334,167 6,678 4,483,433 979 3,862,568 7 3,420,000 *hg13 2002 20,285 4,716,001 4,515 3,465,940 488 3,337,874 7 3,378,000 hg15 2003 14 4,210,361 5 3,218,979 3 8,010,000 7 4,219,878 *hg16 2003 13 4,182,522 5 1,461,098 5 8,026,000 8 4,316,878 *hg17 2004 10 974,522 4 1,460,998 5 8,026,000 6 2,930,711 *hg18 2006 10 974,522 4 1,460,998 5 8,026,000 6 2,930,711 *hg19 2009 7 3,400,000 20 3,420,019 6 3,320,000 7 3,520,000 *hg38 2013 48 337,237 61 283,680 9 176,858 90 499,910 CHM13v2 2022 0 0 0 0 0 0 0 0 assembly year ------ chr20----- ------ chr21----- ----- chrX ------ ------ chrY ----- hg1 2000 11,919 7,629,675 471 2,325,440 14,929 15,861,115 1,266 1,562,653 hg2 2000 7,973 8,378,942 702 5,187,686 10,397 41,548,747 1,194 3,057,221 hg4 2000 3,070 8,565,162 21 1,083,592 9,668 48,158,870 343 3,467,364 hg5 2000 983 6,446,777 22 11,083,592 8,665 40,053,263 199 29,761,374 *hg6 2000 967 5,641,164 22 11,083,592 7,381 34,143,310 146 30,506,644 hg7 2001 8 5,709,984 22 11,083,592 4,577 26,265,292 161 29,208,759 hg8 2001 8 5,709,984 22 11,083,592 2,988 22,414,982 7 28,200,000 *hg10 2001 9 3,809,988 52 10,792,471 2,375 10,529,739 6 35,700,000 hg11 2002 6 3,455,826 33 10,792,450 1,688 9,284,472 6 35,700,000 hg12 2002 7 3,420,000 30 10,791,932 1,096 5,286,937 6 35,700,000 *hg13 2002 7 3,378,000 28 10,791,912 926 4,330,290 6 27,700,000 hg15 2003 7 4,219,878 15 13,051,795 37 4,947,502 7 28,200,000 *hg16 2003 8 4,316,878 10 13,051,790 22 4,477,000 9 25,637,000 *hg17 2004 6 2,930,711 11 12,774,217 20 4,430,000 14 32,830,000 *hg18 2006 6 2,930,711 11 12,774,217 16 3,855,000 14 32,120,000 *hg19 2009 7 3,520,000 17 13,023,253 23 4,170,000 18 33,720,000 *hg38 2013 90 499,910 52 6,621,364 34 1,147,866 61 30,812,372 CHM13v2 2022 0 0 0 0 0 0 0 0
The title of the Scientific American Graphic Science visualization is "3,117,275,501 bases, 0 gaps". This total is based on the length of chromosomes 1–22, X and Y in the CHM13v2 assembly and excludes chrM.
The table below shows how the number and size of gaps and sequence length in each chromosome's assembly changed from 2013 to 2022.
human genome assembly -------------------------------------------------------------- CHM13v2 hg38/GRCh38 Mar 2022 Dec 2013 CHM13v2 new ---------------- --------------------------------------------- ----------- chr seq gap seq ------ gap ------ tot 1 248,387,328 0 230,481,012 168 18,475,410 248,956,422 17,906,316 2 242,696,752 0 240,548,228 35 1,645,301 242,193,529 2,148,524 3 201,105,948 0 198,100,135 29 195,424 198,295,559 3,005,813 4 193,574,945 0 189,752,667 18 461,888 190,214,555 3,822,278 5 182,045,439 0 181,265,378 37 272,881 181,538,259 780,061 6 172,126,628 0 170,078,522 16 727,457 170,805,979 2,048,106 7 160,567,428 0 158,970,131 20 375,842 159,345,973 1,597,297 8 146,259,331 0 144,768,136 12 370,500 145,138,636 1,491,195 9 150,617,247 0 121,790,550 46 16,604,167 138,394,717 28,826,697 10 134,758,134 0 133,262,962 80 534,460 133,797,422 1,495,172 11 135,127,769 0 134,533,742 17 552,880 135,086,622 594,027 12 133,324,548 0 133,137,816 30 137,493 133,275,309 186,732 13 113,566,686 0 97,983,125 23 16,381,203 114,364,328 15,583,561 14 101,161,492 0 90,568,149 25 16,475,569 107,043,718 10,593,343 15 99,753,195 0 84,641,325 19 17,349,864 101,991,189 15,111,870 16 96,330,374 0 81,805,943 22 8,532,402 90,338,345 14,524,431 17 84,276,897 0 82,920,204 48 337,237 83,257,441 1,356,693 18 80,542,538 0 80,089,605 61 283,680 80,373,285 452,933 19 61,707,364 0 58,440,758 9 176,858 58,617,616 3,266,606 20 66,210,255 0 63,944,257 90 499,910 64,444,167 2,265,998 21 45,090,682 0 40,088,619 52 6,621,364 46,709,983 5,002,063 22 51,324,926 0 39,159,777 49 11,658,691 50,818,468 12,165,149 X 154,259,566 0 154,893,029 34 1,147,866 156,040,895 -633,463 Y 62,460,029 0 26,415,043 61 30,812,372 57,227,415 36,044,986 ------------- --- ------------- ----- ----------- ------------- ----------- 3,117,275,501 0 2,937,639,113 1,001 150,630,719 3,088,269,832 179,636,388 M 16,569 0 16,568 1 1 16,569 1 ============= === ============= ===== =========== ============= =========== 3,117,292,070 0 2,937,655,681 1,002 150,630,720 3,088,286,401 179,636,389 Un* 111,660,102 240 9,339,602 ============= ===== =========== ============= 3,049,315,783 1,242 159,970,322 3,209,286,105 *chrUn represents 430 unanchored, random and alternate haplotype contigs.
I did not include the mitochondrial chromosome in the total because the visualization does not show this chromosome — it's too tiny (16,569 bases) to show at the resolution used in the graphic (1,000,000 base bins). If we were to include chrM in the total, it would be 3,117,292,070 bases.
The total bases in the “CHM13v2 new” column reflects only the difference in the total assembly size (excluding gaps). As such, it's a rough accounting of new content in the CHM13v2 assembly. The actual number of unique content in CHM13v2 is closer to 213 million bases (CHM13 unique in comparison to GRCh38/hg38 and GRCh37/hg19).
Before the telomere-to-telomere assembly, the most recent assembly was hg38 (Dec 2013). This assembly had 430 contigs representing unanchored, random and alternate haplotype contigs. These totaled 111,660,102 bases with gaps of 9,339,602 bases. This extra content is part of the assembly, in addition to the chromosome assemblies.

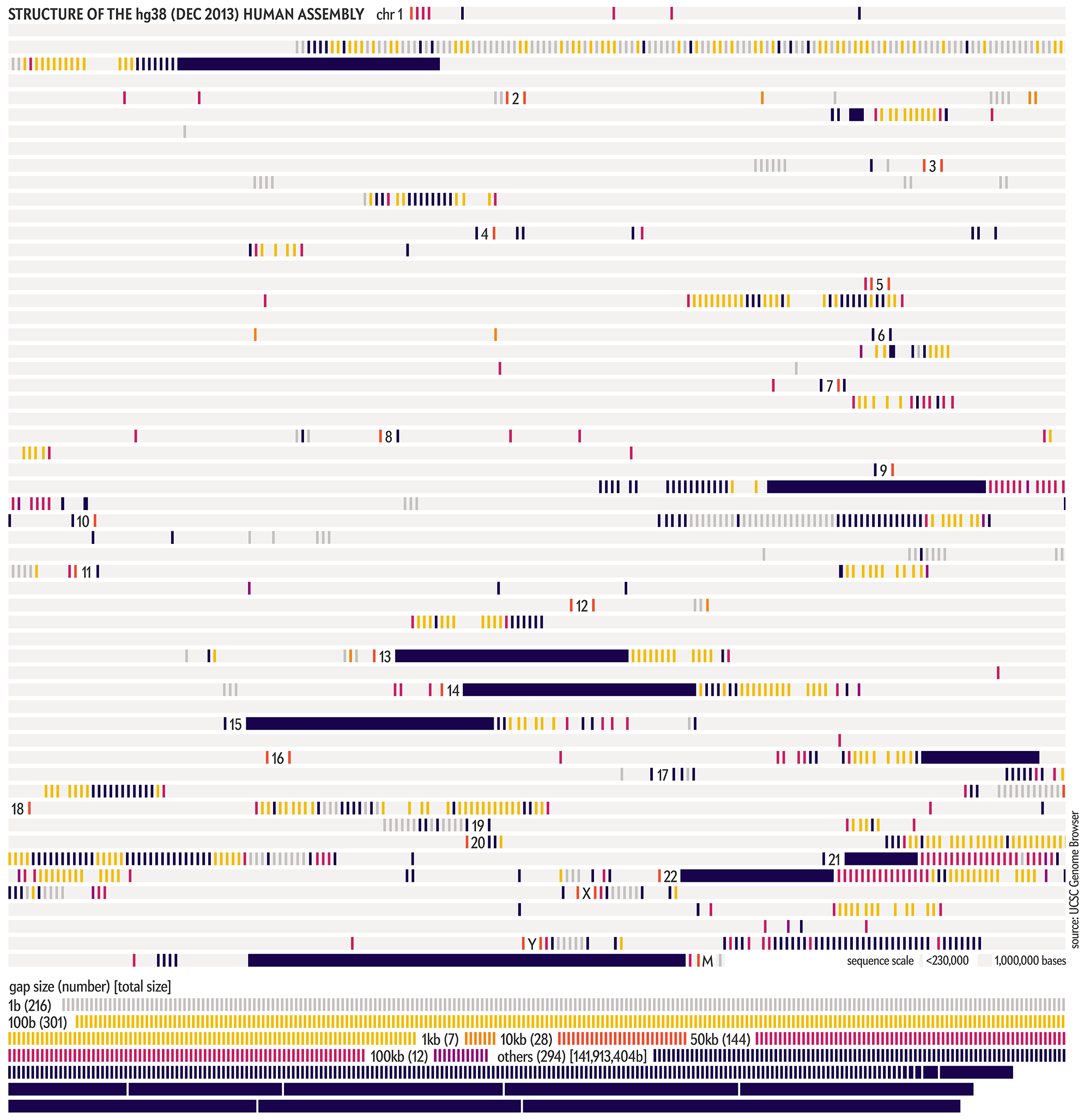
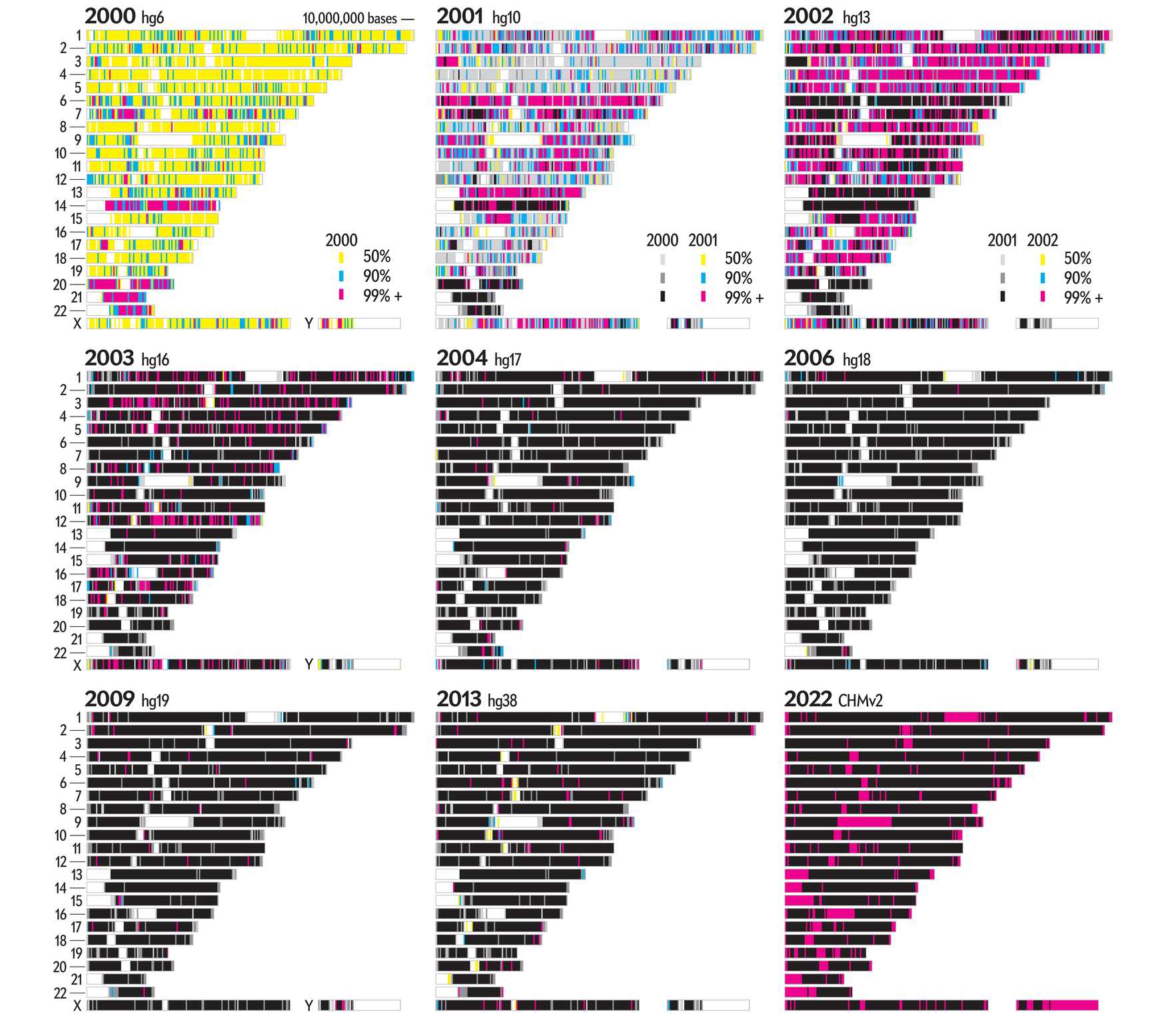
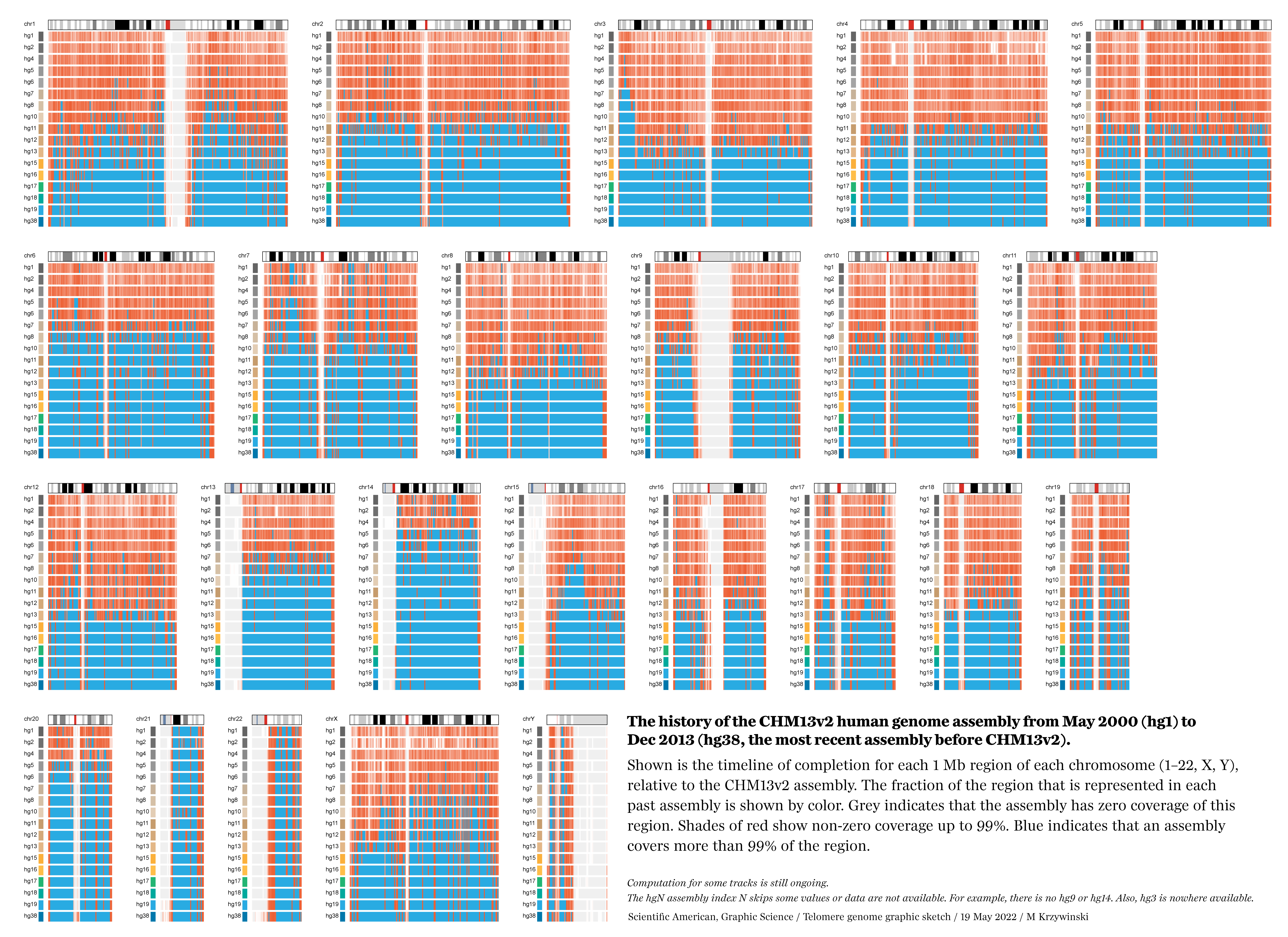
The graphic shows the sequencing history of each of the 22 numbered chromosomes and two sex chromosomes (X, Y) in the human genome. The chromosomes are shown and sized based on their length in the CHM13v2 assembly. Each chromosome is divided into regions of 1,000,000, whose color encodes the degree of completion (50, 90 or 99%+ complete) at the end of a year in which an assembly was available.
These completion cutoffs serve as reasonable milestones. We chose a 99% level of completion as a cutoff instead of 100% to accommodate the differences due to individual variation in the alignments between past assemblies and the CHM13v2 assembly.
WHY 99%? The previous hgN assemblies were constructed from DNA libraries from several individuals, while the CHM13v2 assembly is based on the CHM13hTERT cell line of a single female, with chromosome X sampled from a 51-year-old Harvard University biologist named Leonid Peshkin.
Thus, because we determine year-by-year completion of the assembly by aligning older assemblies to CHM13v2, we always expect a low-level background of differences (at the level of individual variation, which is about 1 base in 1,000), even within regions that are deemed finished. Using a 99% cutoff allows us to sidestep this complication.
Each available assembly from hg1 (May 2000) to hg38 (Dec 2013) (download hg assemblies) was aligned to the CHM13v2 (Mar 2022) assembly as follows.
For a given assembly (e.g. hg38), the assembled sequence of each chromosome was split into chunks of 250,000 bases. For example, splitting hg38 chromosome 1 this way gave 925 chunks (chr1-0000.fa to chr1-995.fa), discarding any chunks that were entirely composed of unknown bases (N's, i.e. the chunk fell in a gap), such as chr-0520.fa.
Each chunk was aligned to its homologous chromsome in the CHM13v2 assembly (e.g. chunks from hg38 chromosome 1 were aligned only to chromosome 1) using blat, a rapid sequence aligner, with the settings shown below and using over-occurring 12-mers via -ooc.
# alignment of hg38 chr1 to CHM13v2 chr1
blat chm13v2/chr1.2bit hg38/chr1.split.2bit -ooc=chm13/chr1.12.ooc
-t=dna -q=dna
-tileSize=12
-minMatch=4 -minScore=100 -minIdentity=98
where hg38/chr1.2bit is the collection of all 250,000 base chunks from hg38 chromosome 1 in 2-bit format. The output of the alignment is in PSL format and provides the mapping between the query Q (e.g., hg38 chromosome 1) and target T (e.g. CHM13v2 chromosome 1).
For example, here's the alignment of chr1-0995.fa, which spans 45 blocks (i.e. the alignment is gapped with 45 closely-spaced individual alignments).
match mis- rep. N's Q gap Q gap T gap T gap strand Q match match count bases count bases name 196082 93 0 2 25 170 20 2922 + chr1-0995 Q Q Q T T T T block size start end name size start end count 206422 0 196347 chr1 248387328 248188227 248387326 45 blockSizes 300,2357,342,4216,13269,70,5142,27806,14859,36195,12552,11060,14049,19514,22919,2869, 6015,1282,36,16,562,32,59,14,62,26,6,24,6,6,29,30,17,18,5,47,87,66,12,118,6,6,5,61,5, qStarts 0,300,2659,3004,7220,20495,20567,25709,53515,68375,104570,117136,128197,142246,161760, 184685,187647,193663,194945,194981,194997,195571,195610,195674,195688,195750,195777, 195784,195809,195816,195823,195852,195883,195900,195919,195924,195971,196058,196124, 196136,196256,196264,196272,196279,196342, tStarts 248188227,248188529,248190886,248191228,248195446,248208715,248208785,248213988, 248241999,248256858,248293081,248305633,248316693,248330746,248350387,248373306, 248376175,248382190,248383473,248383514,248383569,248384131,248384163,248384252, 248386503,248386566,248386592,248386598,248386622,248386628,248386634,248386664, 248386694,248386712,248386730,248386736,248386838,248386971,248387042,248387125, 248387243,248387249,248387255,248387260,248387321,
Chunks that did not align or for alignments that did not complete were redone with slightly looser settings -tileSize=11 -minMatch=2 -minScore=50 -minIdentity=95.
Finally, for any alignments that were still missing or did not complete the process was repeated by chunking the 250,000 base regions into 5,000 base regions and aligning these with the tighter alignment settings shown above (-tileSize=12 ...) but without using over-occurring 12-mers.
For each chunk, alignments were processed as follows.
First, for each alignment a list of intervals on the query and target was created. Second, a running union of target intervals was initialized to the null set. Third, iterating over alignments in decreasing order of total match length (longer alignments first), any alignment whose query interval did not overlap a previously accepted alignment for this region had its target intervals added to the running union.
Once all alignments for a chromosome were processed this way (e.g. hg38 chr1 vs CHM13v2 chr1), the coverage by alignment target intervals for each 50,000 base region of the target chromosome (e.g. CHM13v2 chr1) was calculated. For example, here is the coverage by hg38 on the first several 50,000 regions of CHM13v2 chr1. The last two columns are the key quantities: total bases (e.g. 44,889 bases) in this region that aligned and the fraction of the region (44,889 / 50,000 = 0.89778) that this represents.
hg38 chr1 248387328 0 49999 50000 44889 0.89778 hg38 chr1 248387328 50000 99999 50000 44741 0.89482 hg38 chr1 248387328 100000 149999 50000 49657 0.99314 hg38 chr1 248387328 150000 199999 50000 50000 1 hg38 chr1 248387328 200000 249999 50000 47517 0.95034 hg38 chr1 248387328 250000 299999 50000 49989 0.99978 hg38 chr1 248387328 300000 349999 50000 48838 0.97676 hg38 chr1 248387328 350000 399999 50000 49928 0.99856 ...
Once the coverage of each 50,000 base region of the CHM13v2 assembly by all past assemblies were calculated in this way, the first year in which the region reached 50%, 90% and 99%+ was identified. For example, for the region 100,000,000–100,999,999 on chromosome 1, we have 50% completion in 2000, 90% in 2001 and 99%+ in 2002.
year 2000 hg 6 chr 1 100 100000000 100999999 0 0.5 + year 2001 hg 10 chr 1 100 100000000 100999999 0.5 0.90 + year 2002 hg 13 chr 1 100 100000000 100999999 0.90 0.99 + year 2003 hg 16 chr 1 100 100000000 100999999 0.99 0.99 2002 year 2004 hg 17 chr 1 100 100000000 100999999 0.99 0.99 2002 year 2006 hg 18 chr 1 100 100000000 100999999 0.99 0.99 2002 year 2009 hg 19 chr 1 100 100000000 100999999 0.99 0.99 2002 year 2013 hg 38 chr 1 100 100000000 100999999 0.99 0.99 2002 year 2022 hg 99 chr 1 100 100000000 100999999 0.99 0.99 2002but for the region 246,000,000–246,999,999, there was no coverage in 2000, 50% in 2001, reaching 90% in 2003 and finally 99%+ only in 2022.
year 2000 hg 6 chr 1 246 246000000 246999999 0 0 year 2001 hg 10 chr 1 246 246000000 246999999 0 0.5 + year 2002 hg 13 chr 1 246 246000000 246999999 0.5 0.5 2001 year 2003 hg 16 chr 1 246 246000000 246999999 0.5 0.90 + year 2004 hg 17 chr 1 246 246000000 246999999 0.90 0.90 2003 year 2006 hg 18 chr 1 246 246000000 246999999 0.90 0.90 2003 year 2009 hg 19 chr 1 246 246000000 246999999 0.90 0.90 2003 year 2013 hg 38 chr 1 246 246000000 246999999 0.90 0.90 2003 year 2022 hg 99 chr 1 246 246000000 246999999 0.90 0.99 +
The exact fraction of 50%, 90% and 99%+ alignment coverage is a function of the size of the window over which coverage is determined (I use 50,000 bases).
You can download the full human genome assembly history file here.
In this table I show the fraction of the assembly that was added to a given level of completion for each year. For example, the hg38 (Dec 2013) assembly added 58,387,328 bases to 99%+ completed regions.
level of completion relative to CHM13v2 ------------------------------------------------------------------------------- 50% 90% 99%+ ------------------- ------------------- ------------------- year seq % seq % seq % ---- ------------- ----- ------------- ----- ------------- ----- 2000 1,877,491,287 60.23 532,577,100 17.08 250,000,000 8.02 2001 104,027,534 3.34 494,259,566 15.86 645,000,000 20.69 2002 32,031,701 1.03 290,698,294 9.33 1,213,000,000 38.91 2003 13,460,029 0.43 62,635,144 2.01 347,000,000 11.13 2004 3,000,000 0.10 17,000,000 0.55 46,000,000 1.48 2006 3,000,000 0.10 9,084,080 0.29 10,000,000 0.32 2009 1,276,897 0.04 6,324,548 0.20 33,707,364 1.08 2013 23,000,000 0.74 12,460,830 0.40 58,387,328 1.87 2022 514,180,809 16.49
To correctly interpret this table (and the one below), it's important that you understand how the level of completion of each assembly was determined.
In this table I show the fraction of each assembly that had a given level of completion . For example, the hg38 (Dec 2013) assembly had 2,569,094,692 bases in 99%+ regions, which represented about 82% of the CHM13v2 assembly length.
level of completion relative to CHM13v2 ------------------------------------------------------------------------------- 0% 50% 90% 99%+ ------------------- ------------------- ------------------- ------------------- year seq % seq % seq % seq % ---- ------------- ----- ------------- ----- ------------- ----- ------------- ----- 2000 457,207,114 14.67 1,877,491,287 60.23 532,577,100 17.08 250,000,000 8.02 2001 358,920,014 11.51 1,157,518,821 37.13 731,836,666 23.48 869,000,000 27.88 2002 276,888,313 8.88 250,852,228 8.05 499,534,960 16.02 2,090,000,000 67.05 2003 261,382,845 8.38 124,722,552 4.00 298,170,104 9.57 2,433,000,000 78.05 2004 259,382,845 8.32 101,722,552 3.26 282,170,104 9.05 2,474,000,000 79.36 2006 255,382,845 8.19 95,638,472 3.07 279,254,184 8.96 2,487,000,000 79.78 2009 254,105,948 8.15 94,590,821 3.03 249,871,368 8.02 2,518,707,364 80.80 2013 230,105,948 7.38 96,129,991 3.08 221,944,870 7.12 2,569,094,692 82.41 2022 3,117,275,501 100.0
 buy artwork
buy artwork

All the images here are available as posters.
The final graphic had the genome divided into 1,000,000 base regions. At this resolution, each region is about 0.6 pt wide, which is a reasonable balance of detail and legibility.
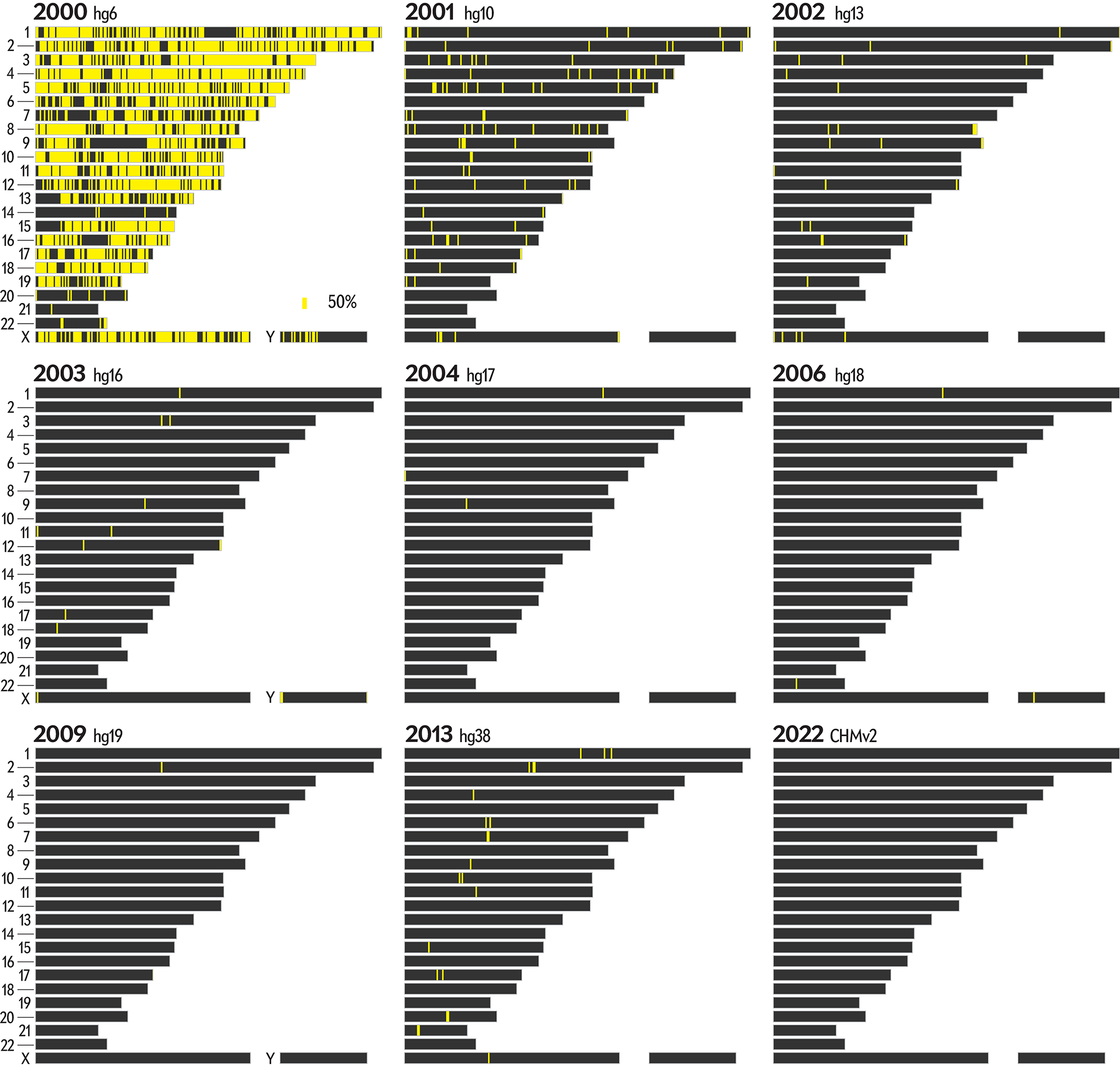
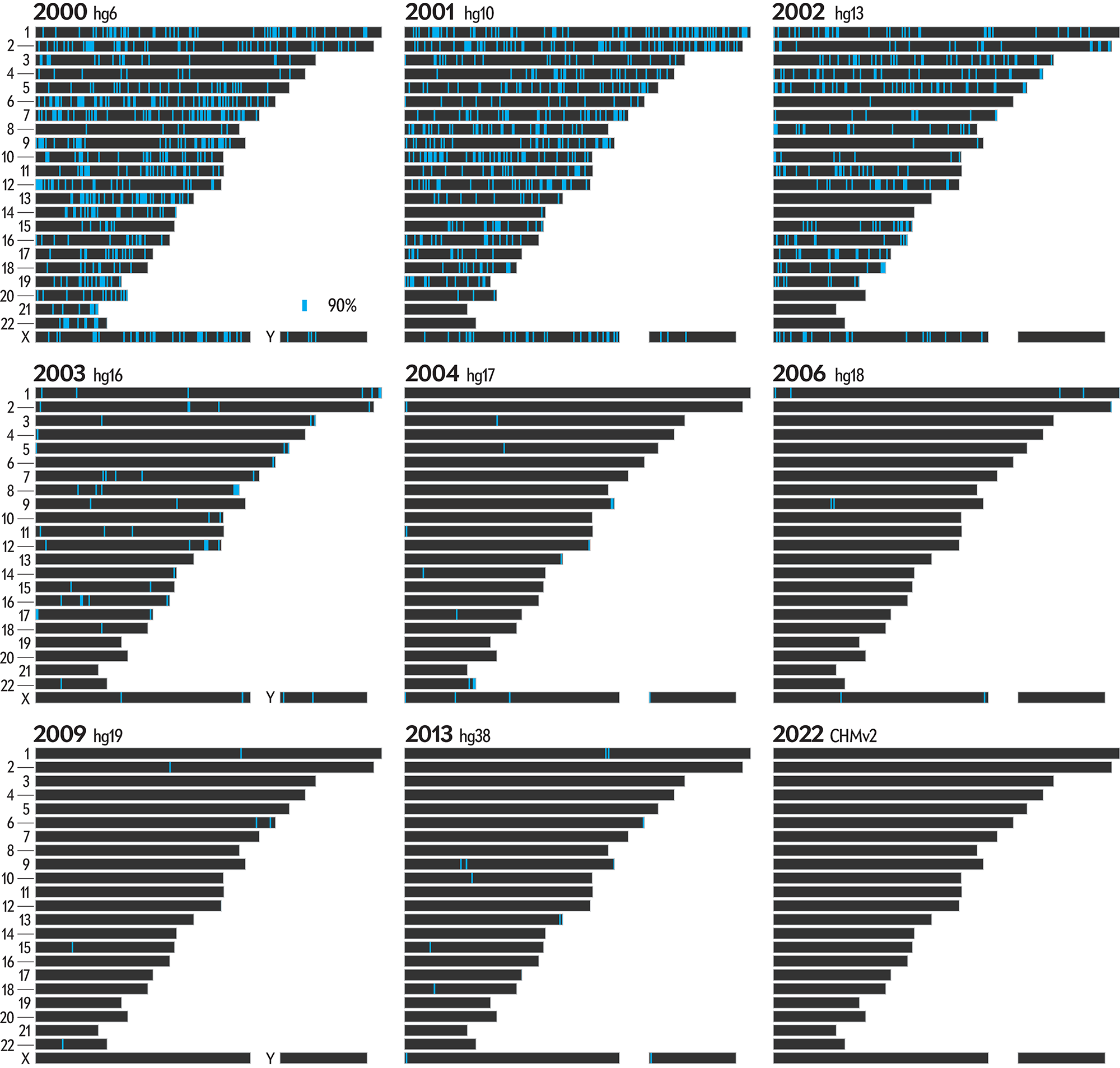
There's a lot going on in this graphic. Below, I show only the regions that flipped to a specific completion level in a year.
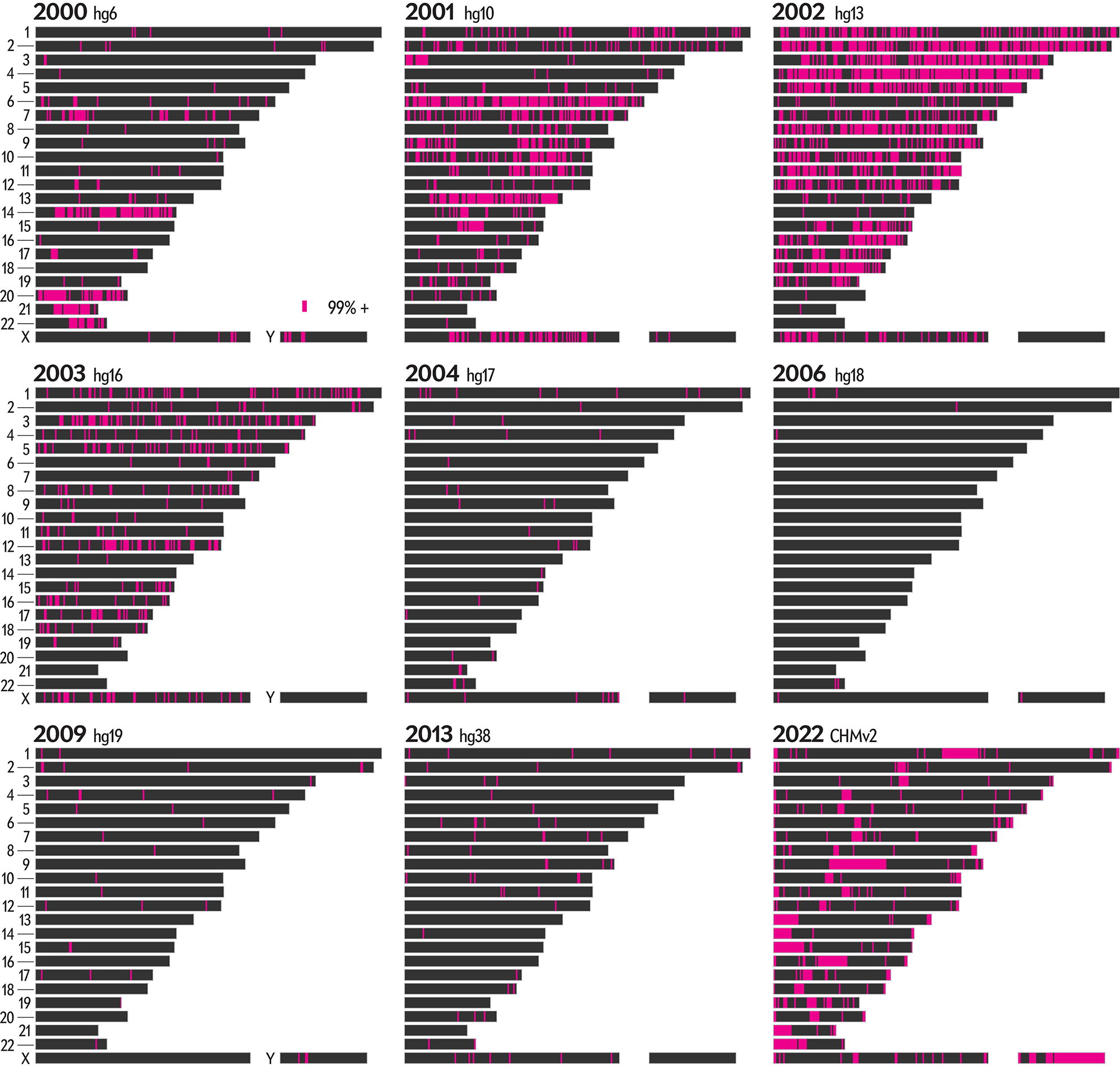
More detailed profiles are possible — here I show the graphic using 250,000 base regions — their detail is hard to discern on the printed page.
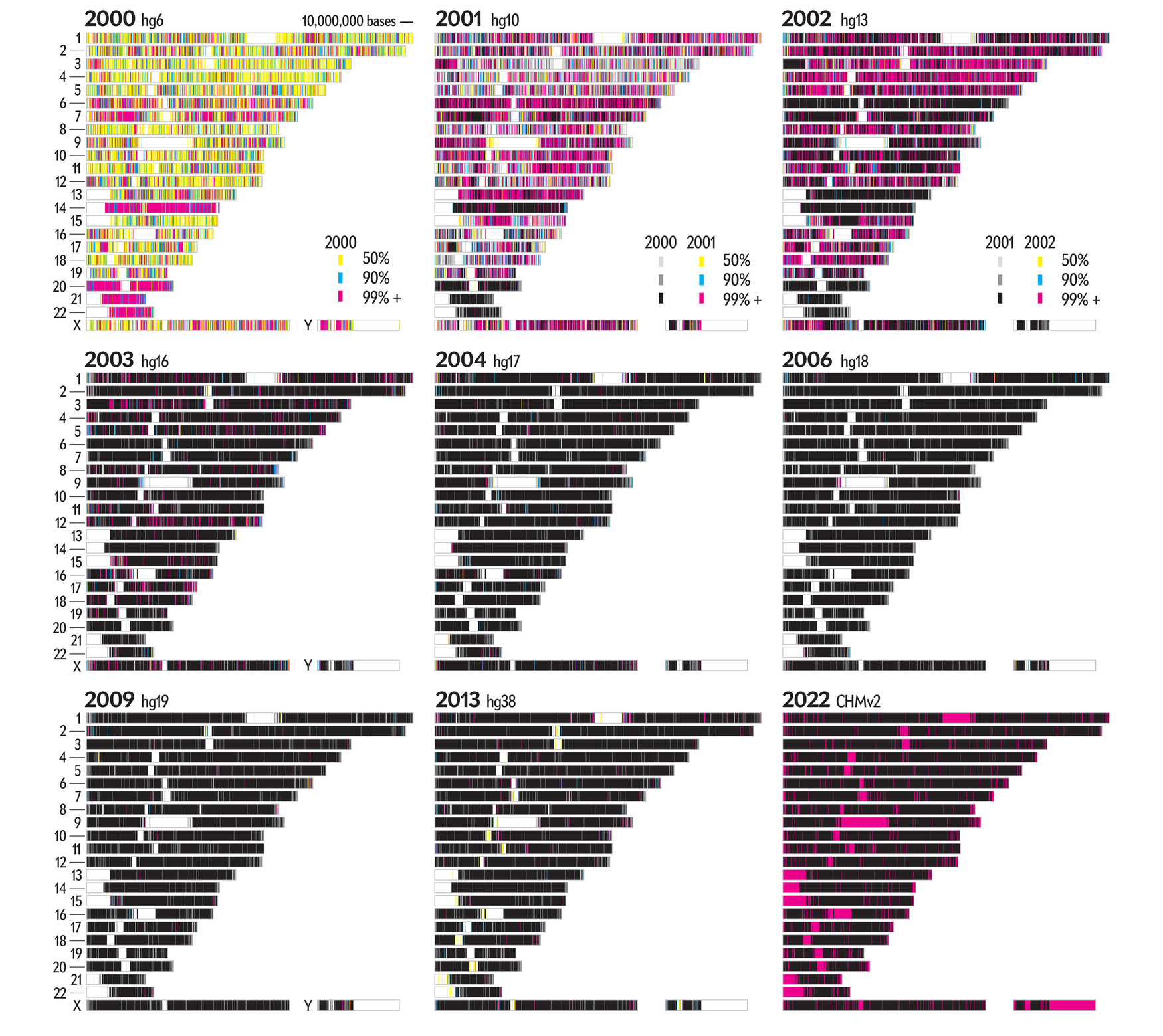
Less detailed profiles provide a good overview — here I show the graphic using 5,000,000 base regions.
While the final graphic shows a snapshot of the genome for each year in which an assembly was available, it's possible to look at the timeline of all the assemblies from hg1 (May 2000) to hg38 (Dec 2013) (assembly release dates). For example, here is a detailed look at progress of completion of chromosome 1 in regions of 1,000,000 bases, with blue indicating 99%+ completion.
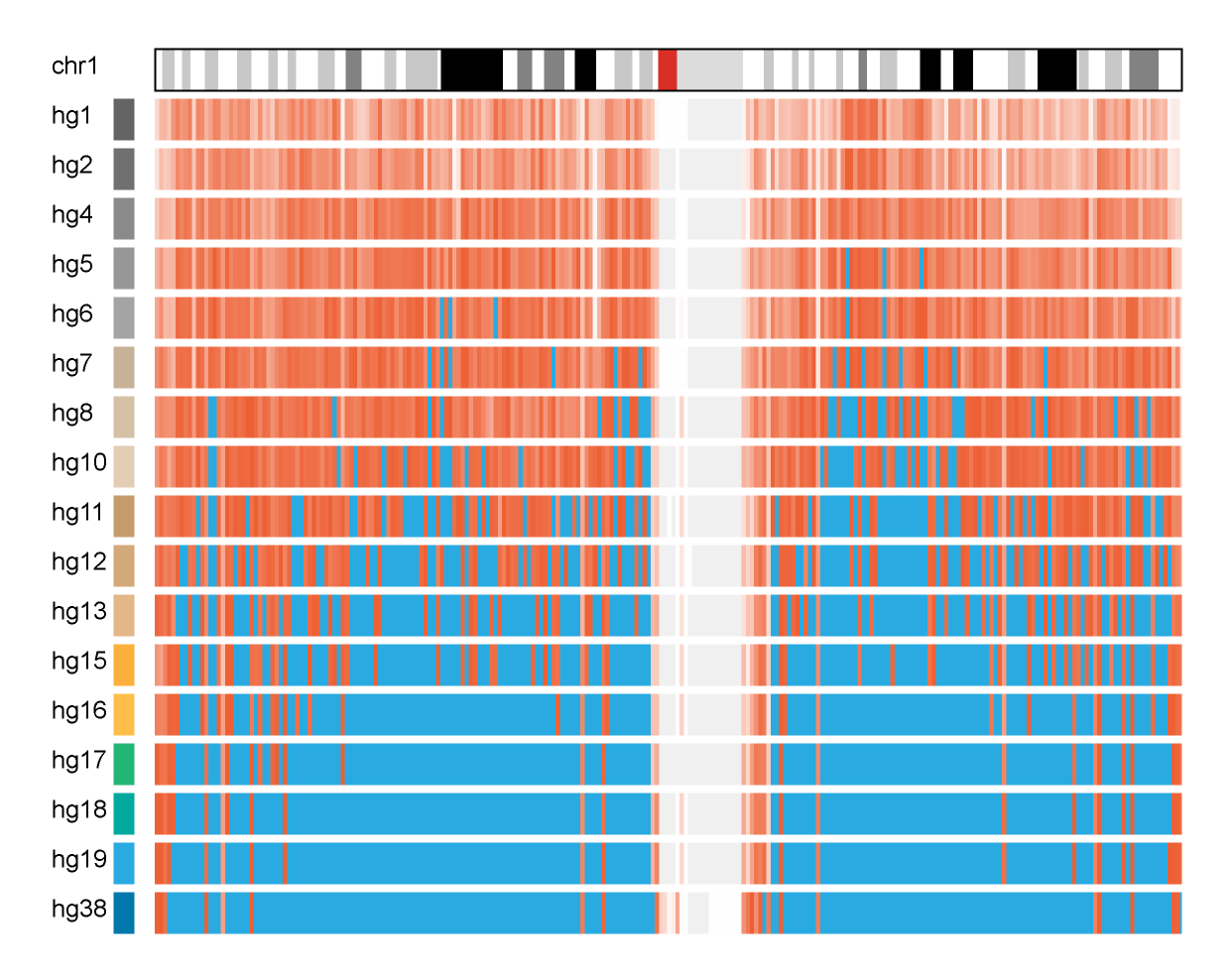
And now drawing all the chromosomes,
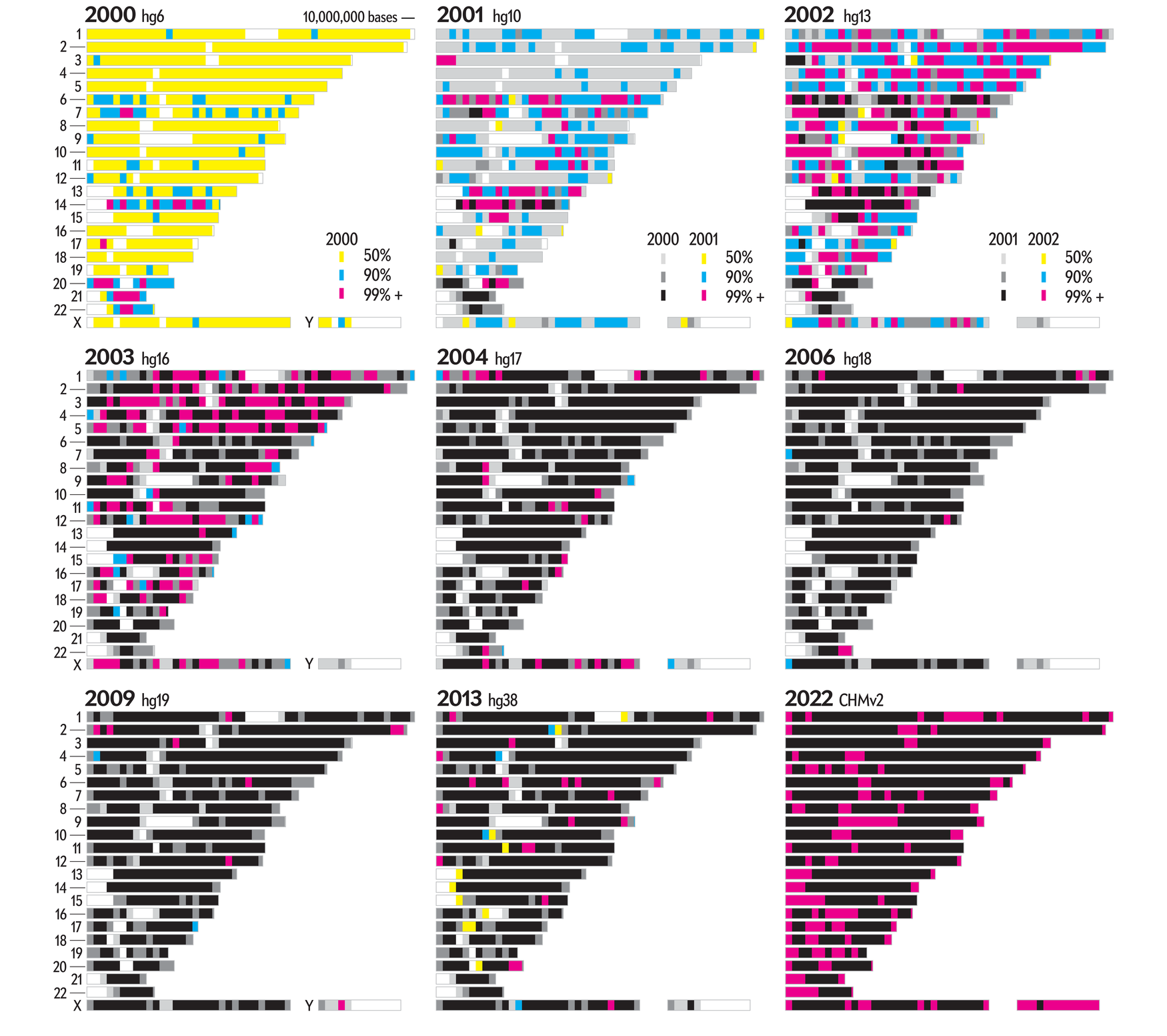
One of the earlier designs for the graphic had the entire genome shown at a higher resolution (50,000 bases per region) with color encoding the assembly in which the region first reached 99%+ completion.
Though it showed promise and definitely showed more data, we thought that this graphic, ultimately, answered fewer interesting questions. But it does make for a pretty poster.
For example, only one level of completion is possible to encode (unless we use textures — yuck) and it's not easy to quickly tell how much activity occurred in any given year. It's also hard to quickly find the start and end of chromosomes.

And here is a take of the assembly at a glance graphic at lower resolution, using 250,000 base regions.

Propensity score weighting
The needs of the many outweigh the needs of the few. —Mr. Spock (Star Trek II)
This month, we explore a related and powerful technique to address bias: propensity score weighting (PSW), which applies weights to each subject instead of matching (or discarding) them.
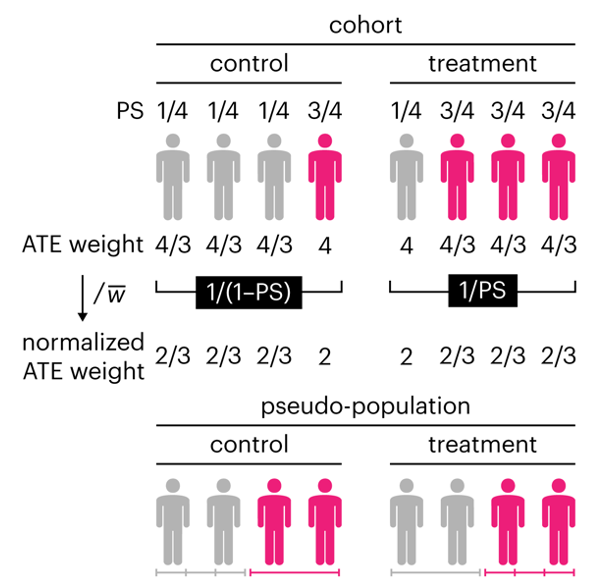
Kurz, C.F., Krzywinski, M. & Altman, N. (2025) Points of significance: Propensity score weighting. Nat. Methods 22:1–3.
Happy 2025 π Day—
TTCAGT: a sequence of digits
Celebrate π Day (March 14th) and sequence digits like its 1999. Let's call some peaks.
Crafting 10 Years of Statistics Explanations: Points of Significance
I don’t have good luck in the match points. —Rafael Nadal, Spanish tennis player
Points of Significance is an ongoing series of short articles about statistics in Nature Methods that started in 2013. Its aim is to provide clear explanations of essential concepts in statistics for a nonspecialist audience. The articles favor heuristic explanations and make extensive use of simulated examples and graphical explanations, while maintaining mathematical rigor.
Topics range from basic, but often misunderstood, such as uncertainty and P-values, to relatively advanced, but often neglected, such as the error-in-variables problem and the curse of dimensionality. More recent articles have focused on timely topics such as modeling of epidemics, machine learning, and neural networks.
In this article, we discuss the evolution of topics and details behind some of the story arcs, our approach to crafting statistical explanations and narratives, and our use of figures and numerical simulations as props for building understanding.
Altman, N. & Krzywinski, M. (2025) Crafting 10 Years of Statistics Explanations: Points of Significance. Annual Review of Statistics and Its Application 12:69–87.
Propensity score matching
I don’t have good luck in the match points. —Rafael Nadal, Spanish tennis player
In many experimental designs, we need to keep in mind the possibility of confounding variables, which may give rise to bias in the estimate of the treatment effect.
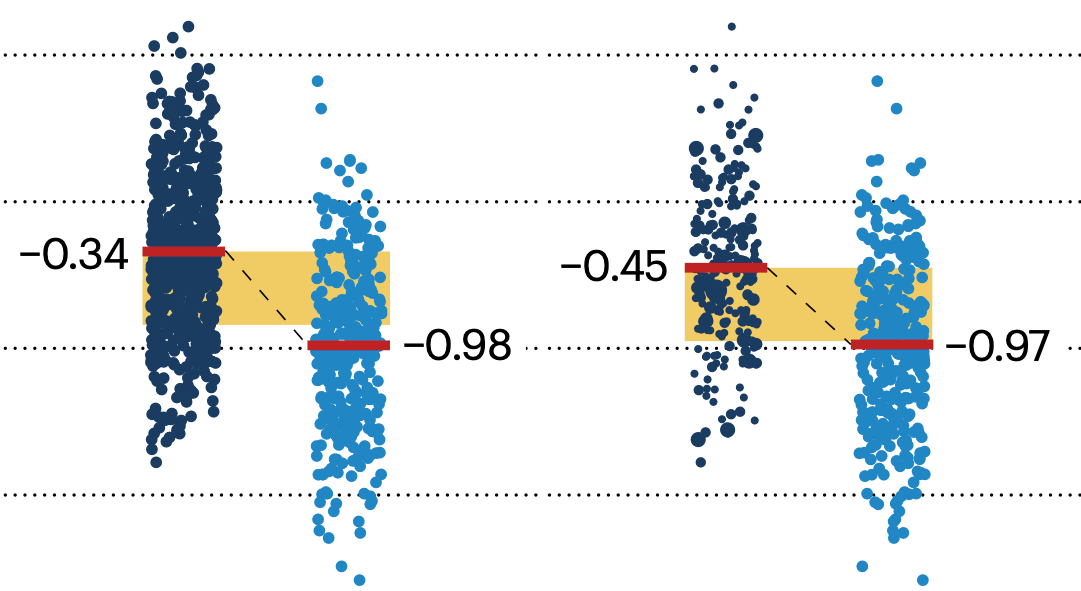
If the control and experimental groups aren't matched (or, roughly, similar enough), this bias can arise.
Sometimes this can be dealt with by randomizing, which on average can balance this effect out. When randomization is not possible, propensity score matching is an excellent strategy to match control and experimental groups.
Kurz, C.F., Krzywinski, M. & Altman, N. (2024) Points of significance: Propensity score matching. Nat. Methods 21:1770–1772.
Understanding p-values and significance
P-values combined with estimates of effect size are used to assess the importance of experimental results. However, their interpretation can be invalidated by selection bias when testing multiple hypotheses, fitting multiple models or even informally selecting results that seem interesting after observing the data.
We offer an introduction to principled uses of p-values (targeted at the non-specialist) and identify questionable practices to be avoided.
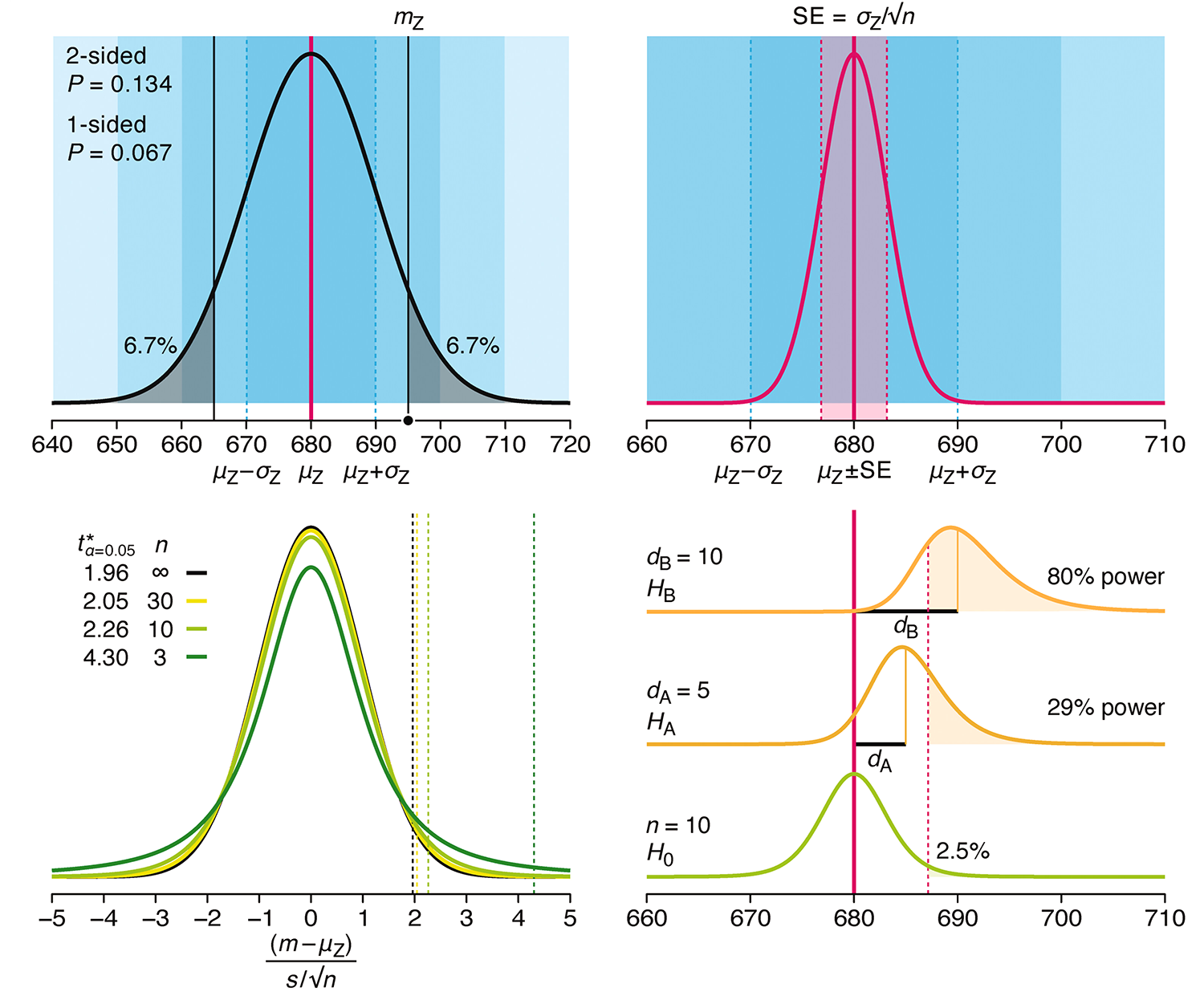
Altman, N. & Krzywinski, M. (2024) Understanding p-values and significance. Laboratory Animals 58:443–446.




